Selenium is a unique and biochemically powerful dietary-essential nutrient that is the key component of 25 human selenoproteins, in which the functions of some are still unknown. Selenium deficiency has much to do with the mutation of viruses and selenium nourishment has a lot to do with protection from virtually all RNA viruses. There’s much more to this relationship than meets the eye. As detailed here, selenium deficiency has now been linked to several viral epidemics.

This can be best understood by following the research of Professor Ethan Will Taylor of the University of North Carolina at Greensboro. Selenium biochemistry research is very difficult to begin with, but even more difficult is research in the relationship between DNA, RNA, and selenium function. Professor Taylor has made several discoveries that significantly add to our understanding of how selenium affects our health. There is no need to wait until the next viral pandemic.
Professor Taylor is a pioneer in this Nobel Prize-worthy research. Research at the Taylor laboratory encompasses areas ranging from structural bioinformatics, molecular modeling, and drug design, to applied molecular biotechnology, nanotechnology, and virology. A major research focus has been on the role of nutritional factors (particularly selenium and dietary antioxidants) in the pathogenesis and progression of HIV/AIDS, Ebola, Zika, COVID-19, and other viral diseases.
Professor Taylor has published more than 80 research articles in these areas. My first interview with Professor Taylor was on AIDS and selenium and was published in our November 1994 issue (1).
Passwater: Congratulations on your recent AJCN publication (2). Please tell our readers what this important research shows.
Taylor:Thanks, and it’s great to be back with you and your readers again. It has become a tradition.
Passwater:That’s because you are the pioneer in viral genomic structure in relation to selenium function. We’ve talked about selenium and various RNA viruses over the years. Later, I am going to ask why RNA viruses as a class (which includes almost all the emerging, zoonotic and pandemic viral threats) are inherently impacted by the selenium status of the host. But, let’s start with your current selenium and COVID-19 study.
Taylor:This new study in AJCN was an international team effort and was made possible in part because the COVID-19 pandemic started in China. That country has geographic regions that range from extremely high to extremely low selenium intakes, which have been very well-documented because of diseases associated with selenium deficiency and viral infections that have been a problem in the low selenium regions in the past. Because of that history, detailed city-by-city data on the amount of selenium in human hair has been published over the years. And of course, the coronavirus case data was being accumulated on a city-by-city basis also, just as we are now doing here in the United States. So, knowing that relatively fine-grained selenium intake data were available, based on past precedents and my own ideas about the mechanisms involved, we were confident that we could test the hypothesis that selenium status might influence the outcome of COVID-19 in these areas.
For our analysis, we used reported cumulative case and outcome data for a snapshot of the progress of the COVID-19 outbreak, as of 2-18-2020. We found that in the city of Enshi, which is renowned for having the highest selenium intakes in China, the cure rate was 3 times as high as that for all the other cities in Hubei Province, where Wuhan is located. In contrast, in Heilongjiang Province, where Keshan is located and extreme selenium deficiency has been known to occur, the death rate was almost 5 times as high as that for all the other Provinces and Municipalities outside of Hubei. Finally, for a set of 17 cities outside of Hubei, using published city-specific data on selenium in human hair, we showed a significant linear correlation with the cure rate for COVID-19. Please see Figure 1.
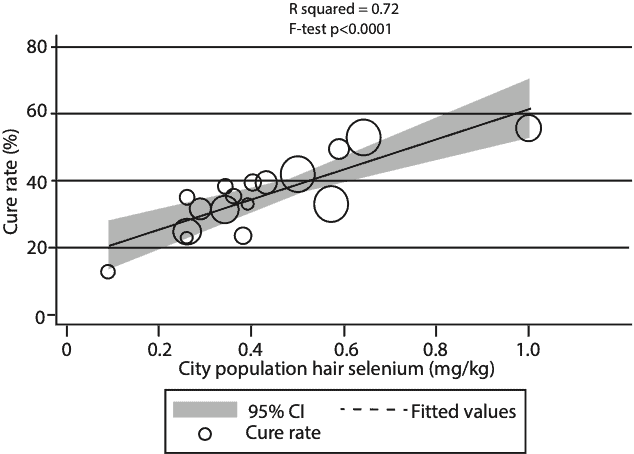
February, 2020 and city population selenium status (hair selenium conentration) analyzed using weighted linear regression (mean ± SD = 35.5 ± 11.1, R2=0.72, F test P < 0.0001). Each data point represents the cure rate, calculated as the number of c
ured patients divided by the number of confirmed cases, expressed as a percentage. The size of the marker is proportional to the number of cases.
(Open acces Amer. J. Clin. Nutr. Ref 2)
The statistical significance on all these observations was very high, p < 0.0001, meaning there was less than a 1 in 10,000 probability that the observations could have occurred by chance. It is one of the more dramatic correlations between selenium status and the outcome of an infectious disease that has ever been published and was based on a fairly large data set. Of course, the way science works, this does not prove that variations in selenium status were the cause of increased survival vs. mortality in SARS-CoV-2 infection, but it is now a very strong hypothesis that selenium is a major influence on the outcome. So that obviously raises the question, what are the mechanisms involved, and why should selenium be so important?
Passwater:Good question. We will come back to it later in the interview. But first, how did this research come about? Why did you want to look at selenium intake and COVID-19 in the first place?
Taylor:From a personal perspective, my interest in this goes back to 2003 and the SARS epidemic, which in retrospect was a practice run for the COVID-19 pandemic. Unfortunately, we did not seem to learn as much as we should have or make proper preparations for the next outbreak of a genetically related virus. Thus, we have the disaster unfolding today.
When the original SARS genomic sequence became available early in 2003, similar to what I had seen in HIV in 1994, I was able to identify 2 programmed ribosomal frameshift sites in the virus, both associated with an in-frame UGA (potential selenocysteine) codon in the overlapping reading frame.
Passwater:Whoa! Professor Taylor, this is where you lose many traditional biochemists. Let me remind our readers that a frameshift mutation is a type of mutation involving the insertion or deletion of a nucleotide in which the number of deleted base pairs is not divisible by three. “Divisible by three” is important because the cell reads a gene in groups of three bases.
Frameshift mutations are among the most deleterious changes to the coding sequence of a protein. They are extremely likely to lead to large-scale changes to polypeptide length and chemical composition, resulting in a non-functional protein that often disrupts the biochemical processes of a cell. But viruses have figured out how to frameshift intentionally in order to make a new variant of an existing protein, and you have discovered sites of this type in various RNA viruses.
A codon is a nucleotide triplet that translates from the genetic code of DNA or mRNA which amino acid will be added next during protein synthesis.
Taylor:Yes, and we later showed that both SARS frameshift sites were functional via in vitro assays. At that time, way back in 2003, after learning of my findings, Dr. Jinsong Zhang (who is the joint first author with me on the new 2020 AJCN paper) contacted me with evidence of selenium abnormalities in the blood of SARS patients. So we joined forces and presented our results in an oral presentation that I gave in July 2003 at the International Science Symposium on SARS in Beijing, right after the SARS outbreak had ended. The Abstract for that presentation is now posted on my ResearchGate site (3). Those results were never published at the time because the SARS outbreak was contained, and with the threat no longer imminent, it was not possible to get funding to pursue the findings. Those old results (which we are now preparing to publish) are still very important, because my genomic analysis of the 2019 SARS-CoV-2 virus shows that the 2 frameshift sites that were in the 2003 virus, along with their overlapping in-frame UGA codons, one of which was in a region with significant protein sequence similarity to the prototypical selenoprotein glutathione peroxidase, are also found in the 2019 SARS-CoV-2 virus. So this realization earlier this year strongly suggested to me a role for selenium in the pathology of COVID-19.
Thus, I was not at all surprised when Dr. Zhang, with whom I had collaborated on a number of selenium studies in the intervening years, contacted me in early February. He asked if I had any such genomic results on the new virus, because he thought we might be able to make a correlation between the COVID-19 mortality data and selenium status. At that point if you just looked at the COVID-19 outcome data for Enshi City, known to have extremely high selenium intakes, versus Hailongjiang province, where selenium intakes are notoriously low, it was pretty clear that we were likely to find a significant correlation. But compiling all the data we needed and finding the best way to analyze and present it took some weeks of back-and-forth between the collaborators. With the leadership of Dr. Margaret Rayman of the University of Surrey and several other UK team members that she brought in, we got the results out, and that’s why you and I are talking today.
Passwater:What other epidemics or outbreaks have been found to be linked to selenium deficiencies?
Taylor:Of course the answer to this question could have been part of that to the previous question, because all of the past precedents for RNA viral diseases in particular being linked to selenium status, both in China and in other locations worldwide, were compelling motivations for us to pursue the study. Many of those precedents were established in China, because of the extensive areas that are selenium deficient, and the fact that there was a lot of subsistence farming back in the mid-twentieth century, making the population more susceptible to a deficiency in local soils.
The first of these historically was of course Keshan disease, a non-obstructive cardiomyopathy that was observed going back to the 1930s but was not linked to selenium until years later, and peaked in the 1960s. It was initially considered to be one of the first human selenium deficiency diseases but later proved to have a viral cofactor, Coxsackievirus. As you and I have discussed in this column in October 1996, Dr. Melinda Beck of UNC Chapel Hill conducted some famous studies in the 1990s, showing that in a mouse model of Keshan disease, an initially benign strain of Coxsackievirus could mutate in selenium deficient mice and become more virulent, capable of causing heart damage in selenium adequate mice. Her group later showed similar findings for influenza A (4). There are a number of studies suggesting a benefit of selenium vs. flu viruses, ranging from a 1986 Russian study showing inhibition of influenza virus in cell culture by sodium selenite, to a new 2020 study in which selenite was shown to block the entry of H1N1 influenza virus into cells; there have also been additional supportive studies in mice and humans.
But focusing back on China, in the very low selenium regions, at one time liver disease and primary liver cancer linked to hepatitis viruses was endemic. This disease was shown to be hugely reduced by the use of selenized table salt, much as we use iodinated salt to add iodine to our diets. Another group of common viral infections throughout Asia are caused by hantavirus infections, which in a 2015 study were shown to be 6 times more frequent in low selenium regions than in selenium adequate regions in China. Hantavirus infection is responsible for “epidemic hemorrhagic fever,” which in a study published by Dr. J.C. Hou in 1997, involving an outbreak in Mongolia about 40 years ago, was treated with a short 9-day course of high-dose sodium selenite, resulting in an 80% decrease in mortality. In the light of the above-mentioned 2015 study showing an association with hantavirus infection and low selenium regions in China, this suggests a clear precedent and rationale for trying sodium selenite versus COVID-19, for which we have shown an even stronger association between selenium status and a viral pathology. So the precedents are numerous, and quite compelling; they are summarized in more detail in a slide presentation that I have posted online (5).
Passwater:We have not yet even mentioned Ebola and HIV, which both have interesting stories that probably deserve to be told. Let’s start with Ebola. Is there evidence of a role for selenium in Ebola pathology and treatment?
Taylor:Ebola has been a very frustrating case for me personally, because I have long believed that selenium should have considerable life-saving benefits in Ebola infection, based on my own work and significant precedents such as the successful use of pharmacological doses of sodium selenite versus epidemic hemorrhagic fever by Dr. Hou many years ago. For example, selenium plays a significant role in blood clotting, and has an anti-clotting effect. Excessive clotting and thrombosis is a primary characteristic of Ebola and at least a secondary characteristic of COVID-19, which has been associated with increased incidence of stroke and other clotting-related vascular disorders.
Prior to 2014, most past Ebola outbreaks tended to progress and burn out so rapidly that there was never time to really convince anyone to look at selenium-related issues in Ebola patients. But in the summer of 2014, I was doing basic genomic research on the new variant of the highly lethal Zaire strain of Ebola that had re-emerged in W. Africa and was threatening to possibly become a global pandemic. I had just made a new discovery about this virus with implications for the role of selenium in many RNA viral infections, including HIV-1. This discovery was eventually published inCurrent Topics in Medicinal Chemistryin 2016 (6). This was an extension and refinement of ideas that I had published years earlier about HIV and Ebola, which was apparently cited as a partial basis for the initiation of a small pilot clinical trial that was started in the Ebola Treatment Unit (ETU) at the ELWA Hospital in Monrovia, Liberia, directed by Dr. Jerry Brown, a Liberian surgeon. He was later on the cover of Time Magazine as one of the Ebola Fighters, who were the Persons of the Year in 2014. But selenium was never mentioned in that article, even though its reportedly successful use was what had contributed to his fame in Liberia.
I learned about the clinical trial from one of the medical directors of the hospital there, and apparently there was such a positive response after about a week of supranutritional doses of sodium selenite that the nurses had insisted stopping the study and giving the selenium to the control group patients as well. Then they ran out, and had great difficulty getting additional selenium into the country because of air travel embargoes. It later turned out that the idea and initial protocol for the trial had come from Mr. Howard Armistead, who was aware of my earlier work on Ebola virus, and also of the extensive evidence for the benefit of selenium in HIV infection. For reasons that remain unclear to me, those results were never reported, except anecdotally in the local African Press. However, much more recently, Dr. Jerry Brown has been quoted online as saying “There is a need for more research on what we did. It wasn’t rocket science, just multivitamins and the selenium that you can buy over the counter” (7). And now there are press reports that Dr. Brown has just taken delivery of substantial quantities of selenium supplements that are being used to treat COVID-19 patients in the 14th Military Hospital in Liberia. So I interpret that as evidence of a lesson learned and confirmation that there was at least some evidence of a clinical benefit of sodium selenite versus Ebola back in 2014. It would not surprise me if some of the earliest reports of a clinical benefit of selenium vs. COVID-19 will emerge from Liberia, as they seem to be open-minded about the potential use of selenium, because of their experience with Ebola.
Passwater:What did you find regarding Zika?
Taylor:For Zika, so far, there is less direct evidence of a benefit from selenium than there is in the case of Ebola infection. However, in 2016, my collaborator Dr. Jan Ruzicka and I published a paper on the WHO Zika Open preprint server, in which we made a very strong case for the involvement of selenium, based on circumstantial evidence, theoretical analysis, and some preliminary experimental data. We proposed that Zika infection could be mimicking a rare genetic disease called Progressive Cerebello-Cerebral Atrophy (PCCA), the primary symptom of which is microcephaly. It turned out that the underlying genetic defect in children born with this disease results in the complete inability to make any of the selenium-containing proteins in the body. So we showed that Zika had the potential to interfere with production of the selenium transport protein selenoprotein P, which could prevent selenium from reaching the brain of infected fetuses, globally blocking selenoprotein synthesis in the brain, and thereby mimic the conditions and symptoms of PCCA. Based on previous work with selenoprotein P knockout mice, there was reason to suspect that prenatal supplementation with sodium selenite, which can cross the blood-brain barrier, might rectify some of the deficits in the synthesis of all the other selenoproteins, and lead to clinical improvements. But we’re still waiting to hear if any clinical investigations have ever explored this possibility. My doctoral student Gabrielle Dailey has been studying the effects of Zika infection on selenoprotein P expression in cell culture, and we will be releasing those results very soon. Stay tuned.
Passwater:Indeed, I will! How do you explain your complex research to others not in your field?
Taylor:To really explain all that research, I need to go into a bit of technical detail about the breakthrough I made in our 2016 CTMC paper about Ebola and HIV. As you know, many years ago I had predicted that HIV-1 could potentially encode selenoproteins, using complex genetic processes such as frameshifting, and what is now called the “recoding” of a stop codon in the genetic code, UGA, to make it serve as a codon for the 21st amino acid, selenocysteine, which is the form of selenium that contributes to the function of the various selenoproteins in the body. This involved what I have called “hidden” genes, because they are encoded overlapping other known genes, lack a “start” codon, and are non-obvious to a conventional genomic analysis.
We later cloned several of these HIV-1 encoded selenoproteins and experimentally demonstrated that they had enzymatic or other functional aspects that we had predicted, such as targeted protein binding. We published our results on one of these in the Proceedings of the National Academy of Sciences in 2000, showing that HIV-1 encoded a glutathione peroxidase gene overlapping its envelope gene. However, there was still considerable skepticism about these findings in the research community because we had to use a non-viral component to express these proteins in mammalian cells. The reprogramming of the UGA codon in selenoproteins requires the presence of a particular RNA hairpin structure in the messenger RNA (mRNA), called a selenocysteine insertion sequence (SECIS element).
Passwater:Selenium is the only trace element that our genes specify to form an amino acid that is then incorporated into proteins. The codon UGA species selenocysteine, the 21st amino acid. Selenocysteine is not an accident and is the keystone compound needed to make several selenium-containing proteins (selenoproteins).
The SECIS element is an RNA element around 60 nucleotides in length that adopts a stem-loop (hairpin) structure. This pattern of nucleotides directs the cell to translate UGA codons as selenocysteines. (UGA is normally a stop codon). Recognition of the UGA triplet in mRNA for specifying selenocysteine requires an RNA signal sequence, referred to as the Selenocysteine Insertion Sequence (SECIS), located in the 3’ untranslated region of eukaryotic selenoprotein mRNAs. SECIS elements are thus a fundamental aspect of messenger RNAs encoding selenoproteins, proteins that include one or more selenocysteine residues.
Taylor:We were never able to identify a functional SECIS in HIV-1 or Ebola, or any other RNA viruses that we suspected encoded selenoproteins. So, we had to include a mammalian SECIS element in our experiments in order to express the viral selenoprotein. During the 2014 Ebola outbreak I finally realized why we couldn’t find a SECIS element in the mRNAs of these viruses. Viruses contain only the barest elements of the machinery of life, primarily what they need to get in and out of cells and to replicate their RNA or DNA; they hijack all the cellular machinery for almost everything else. So I realized that HIV and Ebola were also hijacking SECIS elements, by latching onto the mRNAs of a cellular selenoprotein called thioredoxin reductase (TR).
Passwater:TR is the only enzyme known to catalyze the reduction of thioredoxin and hence is a central component in the thioredoxin system. The thioredoxin system forms reduced disulfide bonds in cells. How did the viruses hijack TR?
Taylor:They did this using a mechanism called antisense, in which regions of two DNA or RNA sequences are the inverse complement of each other, so they can find each other and pair up to make a double helix, similar to the DNA structure first described by Watson and Crick in their famous paper.
Passwater:As you mentioned, a double-stranded DNA molecule is composed of two strands with sequences that are complements of each other. To help molecular biologists specifically identify each strand individually, the two strands are usually differentiated as the “sense” strand and the “antisense” strand. They sometimes use the terms coding strand and template strand.
Taylor:RNAs targeting other RNAs in particular has proven to be an extremely important way in which genes and protein synthesis are regulated in cells. One of the consequences of an antisense interaction is that it can gum up the works for protein synthesis, which uses mRNA as a template. A consequence of the viral hijacking of the TR mRNA would also be a decrease in the synthesis of the TR protein. Since TR is an important antioxidant protein throughout the body, this viral hijacking process could contribute to oxidant-induced cell death and various aspects of viral pathology.
Passwater:Our first interview was published in November 1994 on AIDS and selenium (1). What are some of the newer developments regarding the role of selenium in HIV/AIDS?
Taylor:When I first proposed that HIV might encode multiple selenoprotein modules, it was a pretty radical concept, and still is to some people, apparently. Considering that only a handful of selenoproteins were known at the time, the idea that a virus would encode one or more was unprecedented. Since then, several much larger DNA viruses were found to encode full length glutathione peroxidase genes, showing that even viruses might need antioxidant protection from time to time. In the ensuing decades, there have been dozens of studies showing progressive selenium deficits that correlate with AIDS disease progression, and a number of compelling studies showing that serum selenium status is highly predictive of mortality in HIV-infected cohorts. By now, there have been at least four well-designed clinical trials that demonstrated some type of benefit from selenium supplementation in HIV+ people, either alone or as part of a multivitamin. Dr. Marianna Baum has been an investigator in much of that work (9).
My research group is focused on trying to provide experimental evidence for the antisense targeting of TR and the SECIS hijacking mechanism that we proposed for HIV-1 and Ebola in 2016. We have been able to demonstrate selenium-dependent read-through of the UGA codon that normally serves as the stop codon for the HIV-1 nef gene, which I explicitly predicted in 1994. We have just submitted a paper on that, which also shows that the process involves TR1, the target of the HIV-1 nef antisense interaction. We have similar results as yet unpublished showing read-through of the UGA stop codon of the Ebola nucleoprotein gene. Some of those results can be found in a poster from a 2015 Paris meeting on Ebola that is available online (10).
So I think the story of the role of selenium in HIV/AIDS is still unfolding and the true depth of the interaction between viruses like HIV-1 and human selenium biochemistry is only beginning to be appreciated.
Passwater:As I have often said before, your research is Nobel Prize-worthy. It’s time for traditional biochemists to embrace the genetics of your research. What is your current research suggesting, particularly regarding COVID-19?
Taylor:After trying to explain my findings about selenium and viruses over the years, and usually finding it necessary to invoke some very complex molecular biology (some of which I have mentioned earlier here), I have finally reached a more fundamental and easy-to-understand explanation of why so many RNA viruses in particular have proven to be more virulent under selenium deficiency and/or show decreased pathogenicity in people with higher selenium status.
The answer lies simply in their nature as RNA viruses. The biosynthesis of DNA inherently competes with RNA synthesis, because it depends on the reduction of ribonucleotides (RNA building blocks) to 2’-deoxyribonucleotides, which are then used to make DNA. This means that RNA viruses can increase viral RNA production by partially blocking the synthesis of DNA. And there are several ways they could try to do that, most of which may be targeted to varying extent by different RNA viruses. But one of the best ways to slow DNA synthesis is one that involves selenium, which I will now explain.
As an essential element in thioredoxin reductase (TR), selenium is a component of the thioredoxin system, which serves as an electron donor for the reduction of ribose to deoxyribose. TR serves to sustain the thioredoxin redox cycle, making it a perfect antisense target for an RNA virus to slow down DNA synthesis, so that there will be more RNA to make into new viruses. (Please see figure 2).
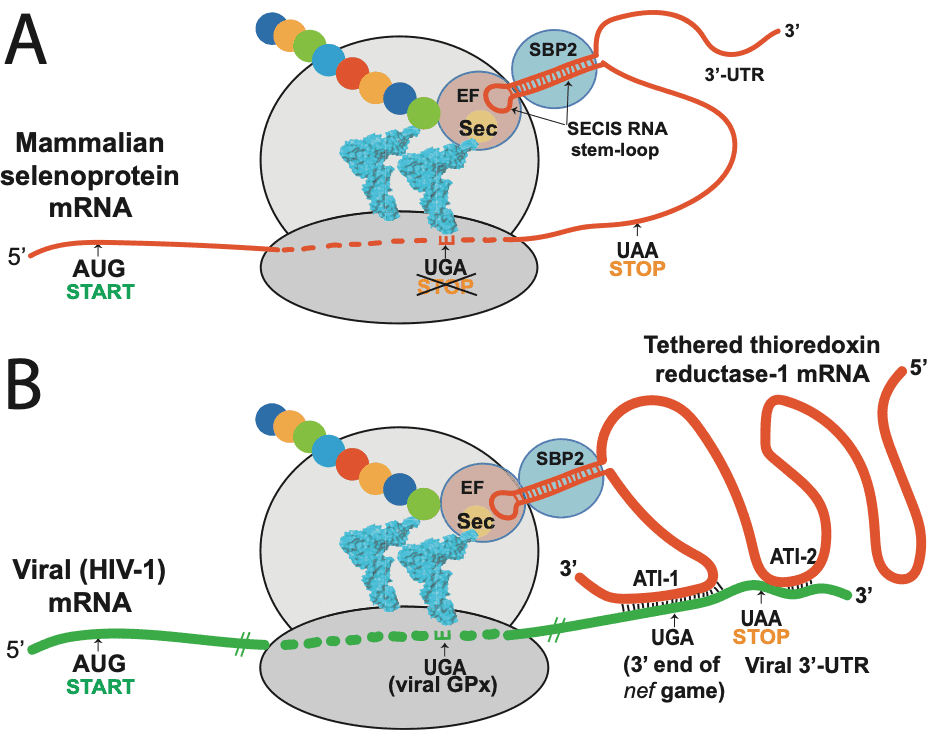
And as you might expect at this point, yes, the SARS-CoV-2 also does this. Like Ebola, it appears to target the TR3 isoform by antisense. We find both anti-human TR3 and anti-Bat TR3 sequences in SARS-CoV-2. One of the anti-TR3 sites is very close by to a frameshift site associated with a conserved UGA codon in the overlapping region, presenting a combination of features that is very similar to the antisense tethering sites we found in Ebola and HIV-1, that we believe are involved in UGA recoding for viral selenoprotein synthesis.
But to end off, I’d like to make a prediction, based on findings from a recent paper in the journal Nature. Gordon et al. studied interactions of most of the SARS-CoV-2 proteins with human proteins, and one of the interactions identified was between the intracellular form of the selenoprotein glutathione peroxidase, GPx1, and a mutant (but not the natural form) of the main viral protease, Mpro. I believe they did not see the interaction of GPx1 with the natural “wild type” protease because the viral enzyme is chopping up the human GPx1. My best current evidence for this is the amino acid sequence of one of the more than 10 sites in SARS-CoV-2 that this viral enzyme cleaves. Using single letter protein code and a slash (/) for the cleavage site, this known protease target is NVATLQ/A. This is very similar to the active site sequence of GPx1, NVASLU/G, where the U stands for selenocysteine, which lines up with a glutamine (Q) in the viral sequence. These two amino acids (U and Q) are similar in size and hydrogen bonding ability. The other “mismatches”, S vs T and G vs A, both differ only slightly, by the size of a methyl group (CH3, a single carbon atom), and are well within the tolerance of this enzyme.
If I am right, this explains a lot about COVID-19 pathogenesis. Not only is the virus using RNA-based antisense mechanisms to cause decreases in the levels of human selenoproteins like TR3, it is probably achieving a similar result by targeting GPx1 for degradation by proteolysis, a protein-based mechanism. In every infected cell, the resulting decreases in the levels of this essential antioxidant enzyme would contribute to increased oxidative stress and risk of cell death, and would be taking place in everyone, regardless of their selenium status. But the effect could vary, from minimal to severe, depending on the viral load and selenium status of the infected person.
Passwater:What’s the bottom line?
Taylor:The bottom line for me is that the association between selenium status and COVID-19 outcomes that we have shown in the AJCN paper is not just simply a reflection of the importance of selenium for a healthy immune system. Something much deeper is going on, and it is not just a concern for people who are selenium deficient. If it is only something that will affect people who are deficient, then why did the people of Enshi City, with a selenium intake far above the recommended daily requirement, have a 3-fold higher recovery rate from COVID-19? What we really need is for some scientists who read this to take it seriously and to try to assess the actual clinical potential of various selenium formulations and to help unravel the complex molecular mechanisms involved.
Passwater:It is frustrating to observe these viral pandemics that do not need to happen. In my humble opinion as a selenium researcher since 1959, you have done much to further our understanding of selenium in human health. Unfortunately, few biochemists are aware of your research in structural bioinformatics and the pathogenesis and progression of HIV/AIDS, Ebola, Zika, COVID-19 and other viral diseases. Hopefully, this publication of your recent simple correlation between dietary selenium and COVID-19 virus survival will attract more researchers to this field. There is no need to wait for the next viral pandemic! Lives can be saved now with the information you have already elucidated.
When I started my selenium laboratory animal research in 1959, there were only a handful of selenium researchers and they were mostly studying animal nutrition. Selenium was not thought to be essential for humans. Neither was vitamin E at that time. I discovered a synergism between selenium and vitamin E in laboratory animals in the early 1960s. I believed then that this could be an unidentified complex tentatively called “Factor-3.” A year later, I found synergism between selenium and vitamin C. Later, it was found that the selenoprotein TR recycled dehydroascorbate to ascorbate (vitamin C) which explains the synergistic action of selenium and vitamin C.
The dietary essentiality of selenium was not established until 1973 when Dr. J. T. Rotruck and his colleagues at the University of Wisconsin determined selenium was a component of glutathione peroxidase (13), Through the years, the functions of many selenoproteins have been identified, but the discoveries described herein may be the most important discovery of all regarding the complex interactions of selenium and human health.
People can improve the survival against many viruses by being adequately nourished with the essential nutrient selenium. The U.S. Institute of Medicine recommends 70 micrograms of selenium per day for men and 55 micrograms per day for women (14).
Remember that selenium is one of the essential nutrients that also can be toxic in large amounts. Do not overdo. One- to two-hundred micrograms of selenium should be optimal. Thank you, Professor Taylor, for once again chatting with us about your research. This pandemic is awfully close to home for most of our readers.
References1. Passwater, R.A. AIDS Discovery Explains the Importance of Selenium: An interview with Will Taylor, Ph.D. Whole Foods Nov. 1994. http://www.drpasswater.com/nutrition_library/taylor_interview1.html 2. Zhang, J., Taylor, E.W., Bennett, K., Saad, R. and Rayman, M.P. Association between regional selenium status and reported outcome of COVID-19 cases in China. Am J Clin Nutr. 2020 Apr 28 : nqaa095. Published online 2020 Apr 28. doi: 10.1093/ajcn/nqaa095 3. www.researchgate.net/lab/Ethan-Will-Taylor-Lab 4. Passwater, R.A. Selenium Against Viruses: More Exciting Research from Dr. Will Taylor. Whole Foods October 1996. http://www.drpasswater.com/nutrition_library/selenium1.html 5. www.researchgate.net/publication/340984525 6. http://www.eurekaselect.com/134956/article 7. https://www.charicomm.org/en/ebola-hero/ 8. Zhao, L., Cox, A.G., Ruzicka, J.A., Bhat, A.A., Zhang, W. and Taylor, E.W. Molecular modeling and in vitro activity of an HIV-1-encoded glutathione peroxidase. PNAS June 6, 2000 97 (12) 6356-6361; https://doi.org/10.1073/pnas.97.12.6356 9. Passwater, R.A. Selenium and viral diseases: facts and hypotheses: An Interview with Professor E. Will Taylor. Whole Foods Sep 1997 http://www.drpasswater.com/nutrition_library/selenium2.html 10. www.researchgate.net/publication/340452921 11. http://dx.doi.org/10.2471/BLT.16.182071 12. Patent US 6,090,414 13. Rotruck JT, Pope AL, Ganther HE, Swanson AB, Hafeman DG and Hoekstra WG. Selenium: biochemical role as a component of glutathione peroxidase. Science 1973 Feb 9;179(73):588-90. 14. Food and Nutrition Board, Institute of Medicine, DRI: dietary reference intakes for vitamin C, vitamin E, selenium and carotenoids. Washington, D.C., National Academy Press 2000; 284-324.










